Boston Children’s Hospital
Division of Gastroenterology, Hepatology and Nutrition
Our Research
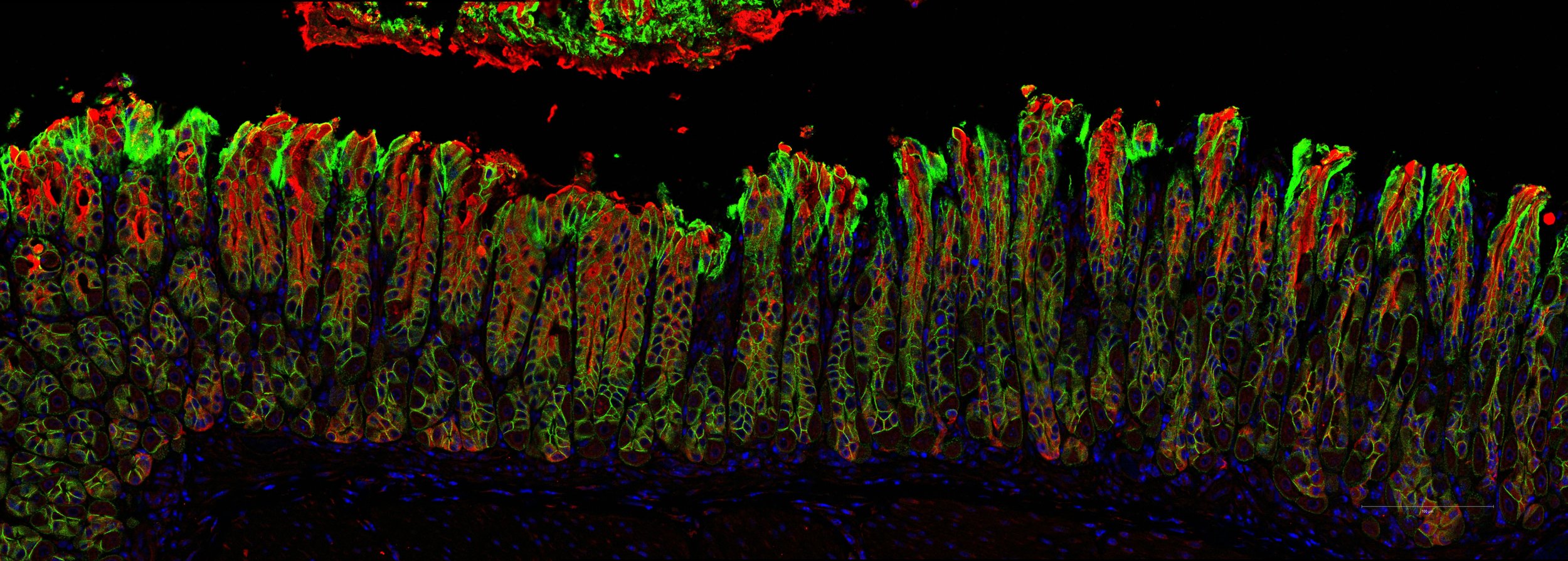
Our laboratory studies the cell and molecular biology of vesicular transport and mechanisms of innate immunity unique to the polarized epithelial cells that line mucosal surfaces. These projects relate to how barrier epithelial cells interact with the luminal and sub-epithelial microenvironment, and to the biology of bacterial pathogenesis and mucosal host defense.
In one project, we have discovered how the enteric bacterial toxin, cholera toxin (CT), breeches the intestinal epithelial barrier and enters host epithelial cells to cause disease. CT and the other AB5-subunit toxins hijack membrane lipids and the cellular and molecular mechanisms of retrograde membrane transport to move from the plasma membrane into the endoplasmic reticulum (ER) of affected cells. Once in the ER, a portion of the toxin, the A1-chain, co-opts components of ERAD (ER-associated degradation) to retro-translocate to the cytosol where it acts enzymatically to cause disease.
Recent studies show that the structure of the ceramide (lipid) domain of the GM1 slycophingolipid receptor plays a decisive role in the trafficking of the toxins backwards in the secretory pathway. Structure-function analysis have elucidated native and non-native ceramide domains that may have clinical applications.
In other studies, we found that upon entry into the endoplasmic reticulum (ER) of host cells, a portion of these toxins activate the unfolded-protein sensor IRE1α. And we discovered the structural basis for unfolded protein recognition by IRE1a leading to induction of the unfolded protein response. Our work on IRE1a led to new projects focused on the biology of its evolutionarily evolved paralogue IRE1b.
The lab also studies the cell and molecular biology of transcytosis by the MHC Class I-like IgG receptor FcRn. FcRn transports IgG across mucosal surfaces where it may function in immune surveillance and host defense. The transcytotic pathway across barrier epithelial cells provides a critical link between the outside (microbial) and inside (sterile) worlds of the gut (and other mucosal surfaces lining the respiratory and genitourinary tracts). Our work has elucidated the itinerary and many genes operating in this pathway.
In a third area of interest, we aim to understand the regulation of ion and water transport in the intestine. These studies address the biology of several ion transporters responsible for secretory diarrhea.
The PARD6B and Apical Endosome research programs
Led By: Stefanie Schmieder
Research Fellow: Julia Ehrmann Student Researcher: Anthony Wang
Research Assistants: Victor Svistunov, Lena Schulz, Tianyang Frank Wang
A new research program emerged from our research on IgG transcytosis by FcRn and the membrane biology of the enterotoxin/glycosphingolipid complexes.
Polarized epithelial cells form the essential barrier against infection at mucosal surfaces. Many pathogens breach this barrier to cause disease, often by co-opting cellular endocytosis mechanisms to enter the cell through the lumenal (apical) cell surface. We recently discovered that loss of the cell polarity gene PARD6B selectively diminishes apical endosome function. We found that in response to epithelial cell entry of certain viruses and bacterial toxins via the apical membrane, PARD6B and aPKC, two components of the PARD6B-aPKC-Cdc42 apical polarity complex undergo rapid proteasome-dependent degradation. Perturbation of apical membrane glycosphingolipids by toxin or virus binding signals to induce the degradation of PARD6B. The loss of PARD6B causes depletion of apical endosome function and renders the cell resistant to further infection from the lumenal cell surface - thus enabling a form of cell-autonomous host defense.
Our lab conducts basic and translational gastroenterology research. This project is specifically interested in how gut microbes might affects hunger, satiety and food choice. The microbiome plays a crucial role in the gut-brain axis serving as the main stream of communication, which influence digestion, the immune system and overall human health. As the prevalence of severe obesity continues to increase, our research is timely and relevant with developing novel therapies to treat obesity. This research uses complementary computational and experimental methods to analyze data, including data science, quantitative analysis, and novel molecular techniques. Most of our work involve creation of bacterial and bacterial supernatant libraries.
Our research is done in collaboration with a team of scientists and clinicians at both Boston Children’s Hospital and Dr. Eric Alm’s lab at MIT who focus on the interaction between gut microbes, the gastroenterology tract and the brain.
Gut-Brain Axis and Microbiome Signaling
Led by: Yanjia Jason Zhang
Research Assistant: Mildred-Maria Reyes
The IBD risk gene INAVA (C1ORF106) research programs
Led by: Wan Nurul Izzati Mohamad
Research Assistant: Victor Svistunov
The lab aims to elucidate how the IBD-risk gene C1ORF106/INAVA acts in human intestinal epithelia to manage environmentally-induced cell stress, inflammation, and the integrity of mucosal surfaces.
C1ORF106, recently named INAVA (Innate Immune Activator (2)), was identified as a risk gene for the chronic inflammatory bowel diseases (IBD) by genome-wide association studies and targeted exome sequencing (3). We became interested in the protein upon finding it to be highly enriched in polarized simple epithelia (4), the cell type that lines the intestine and interfaces most directly with the outside world (the focus of the Lencer lab).
We found that INAVA exhibits dual activities mechanistically linking epithelial barrier function and inflammatory signaling by IL1β. This is driven by INAVA’s signature Domain of Unknown Function DUF3338, which we newly defined as an enhancer of TRAF6-protein ubiquitination. We now know that other ubiquitin-mediated cell-stress pathways are amplified by DUF3338, and that these reactions can broadly affect the proteome via formation of macromolecular cytosolic INAVA-containing puncta. Our work (1) also showed that DUF3338 stably binds the GTP-exchange factor cytohesin-2 (ARNO), in one case blocking the formation of INAVA-puncta (video above) and its effect on ubiquitination, and in another case acting at lateral membranes where the INAVA-ARNO complex regulates cortical F-actin dynamics and epithelial barrier function (1). We thus renamed the domain CUPID for Cytohesin Ubiquitin Protein Inducing Domain (1). Our studies implicate INAVA as a sensor and effector of epithelial proteostasis in barrier function.
This projects focuses on better understanding polarity in the intestinal epithelium and how the epithelium interacts with its environment. Dr. Bugda Gwilt trained with Dr. Gregory Miller are investigating how dietary amines, via TAAR1, could augment intestinal enteropathies.
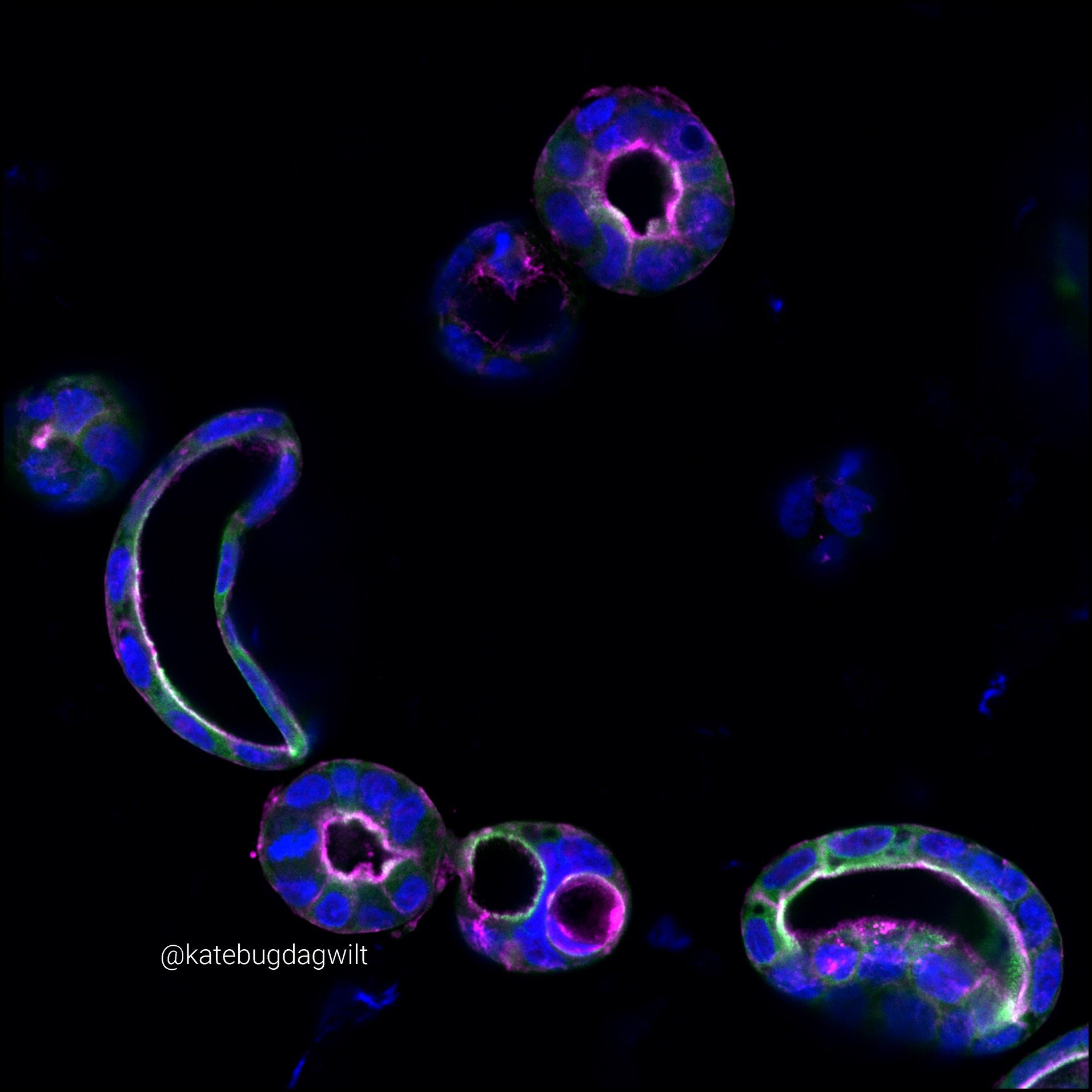
Concurrently, the lab is also studying the mechanisms underlying congenital diarrheal enteropathies using patient derived intestinal organoids. The work focuses on establishing methods for high-content, high-resolution microscopy to investigate the underlying cellular architecture typifying disease, as well as translating existing functional assays into patient derived organoid models to discover novel therapeutics for disease.
Studying the Mechanisms of Intestinal Lumen Formation Using Patient-derived Organoids
Led by: Kate Bugda